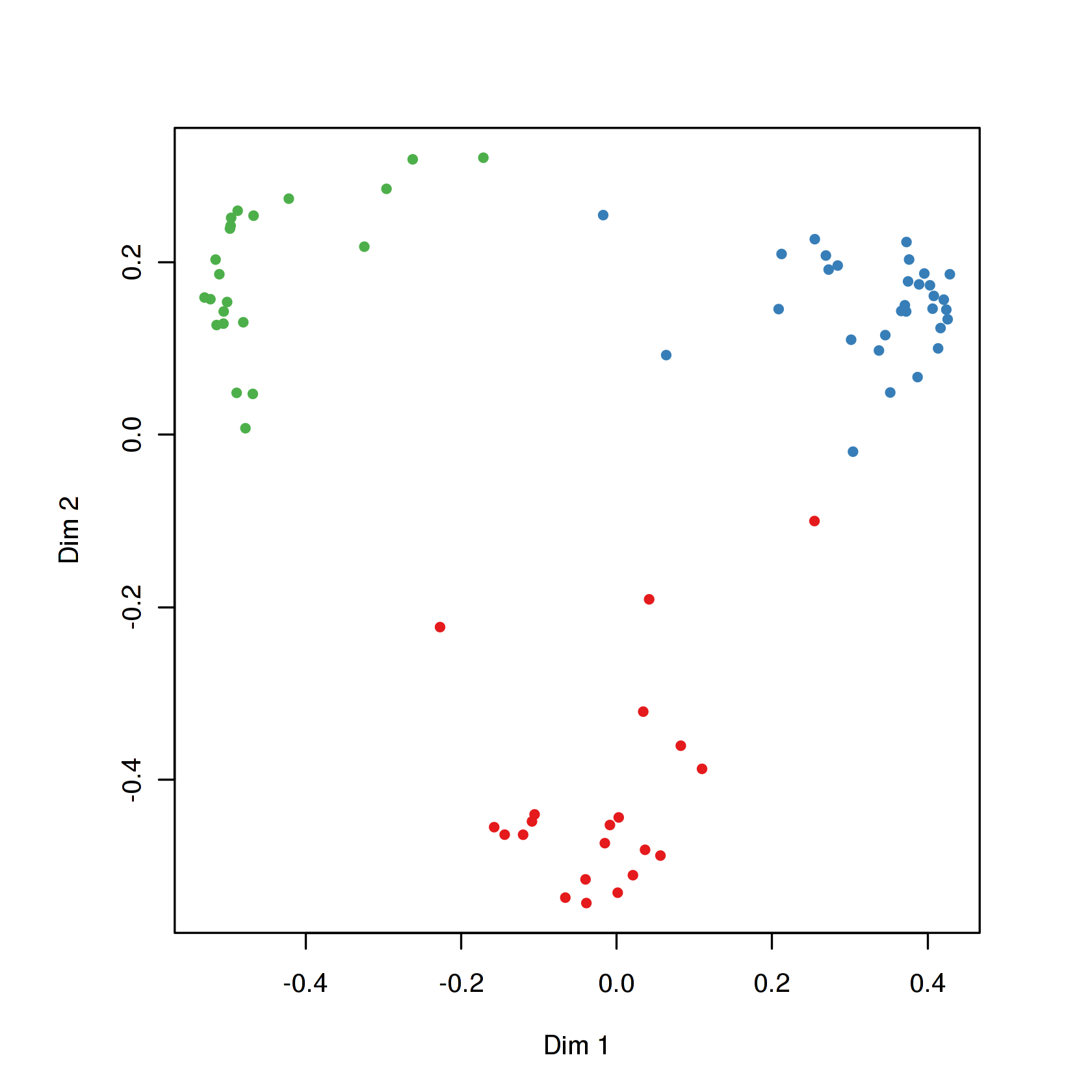
Классификация с обучением
Линейный дискриминантный анализ (LDA)
'data.frame': 74 obs. of 3 variables:
$ Width : int 150 147 144 144 153 140 151 143 144 142 ...
$ Angle : int 15 13 14 16 13 15 14 14 14 15 ...
$ Species: Factor w/ 3 levels "Con","Hei","Hep": 1 1 1 1 1 1 1 1 1 1 ...
- Con
- Con
- Con
- Con
- Con
- Con
- Con
- Con
- Con
- Hei
- Hei
- Hei
- Hei
- Hei
- Hei
- Hei
- Hei
- Hei
- Hei
- Hep
- Hep
- Hep
- Hep
- Hep
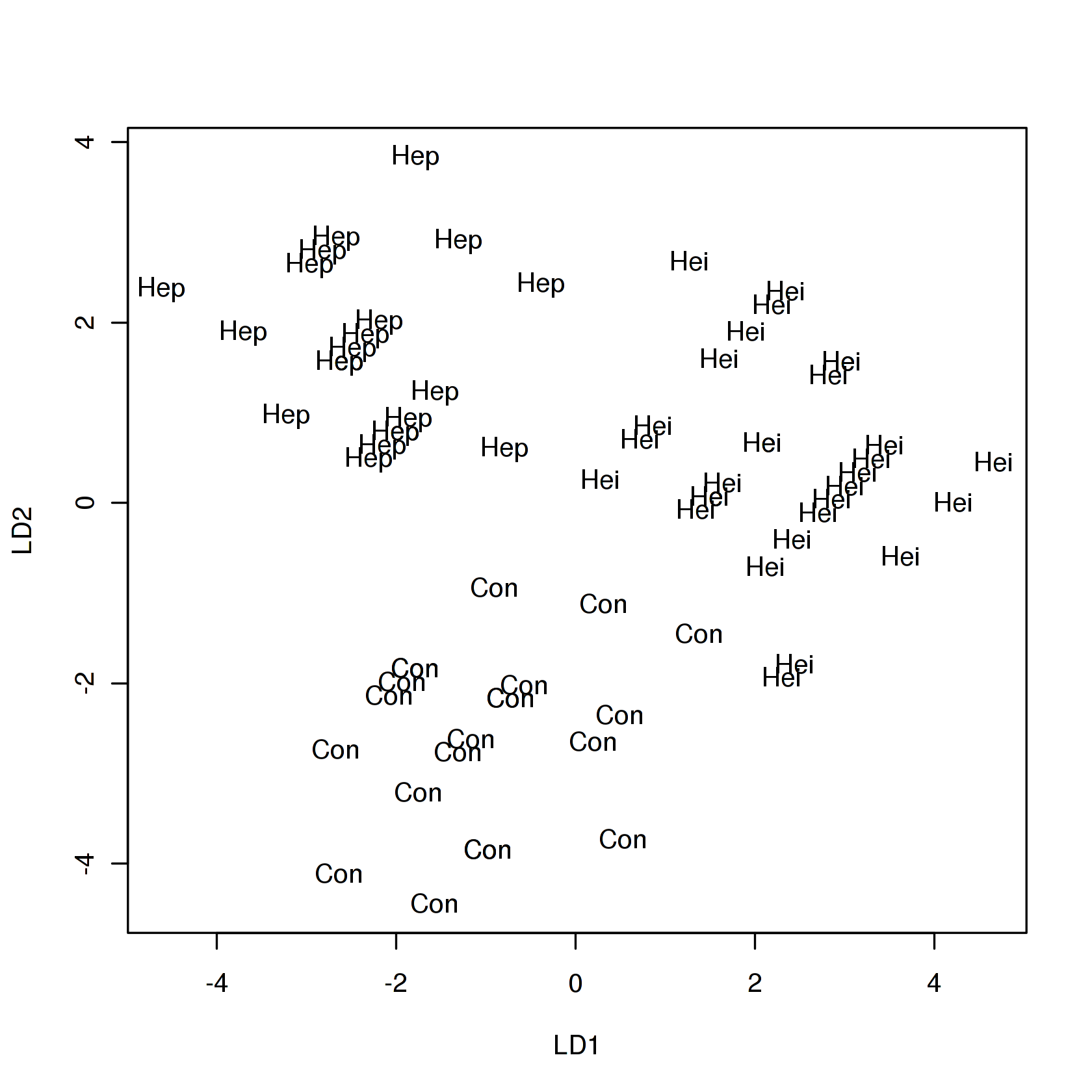
flea.ldap Con Hei Hep
Con 9 0 0
Hei 0 10 0
Hep 0 0 5
Статистическая проверка результатов LDA
Df Wilks approx F num Df den Df Pr(>F)
flea.ldap 2 0.055065 32.615 4 40 4.18e-12 ***
Residuals 21
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Визуализация LDA
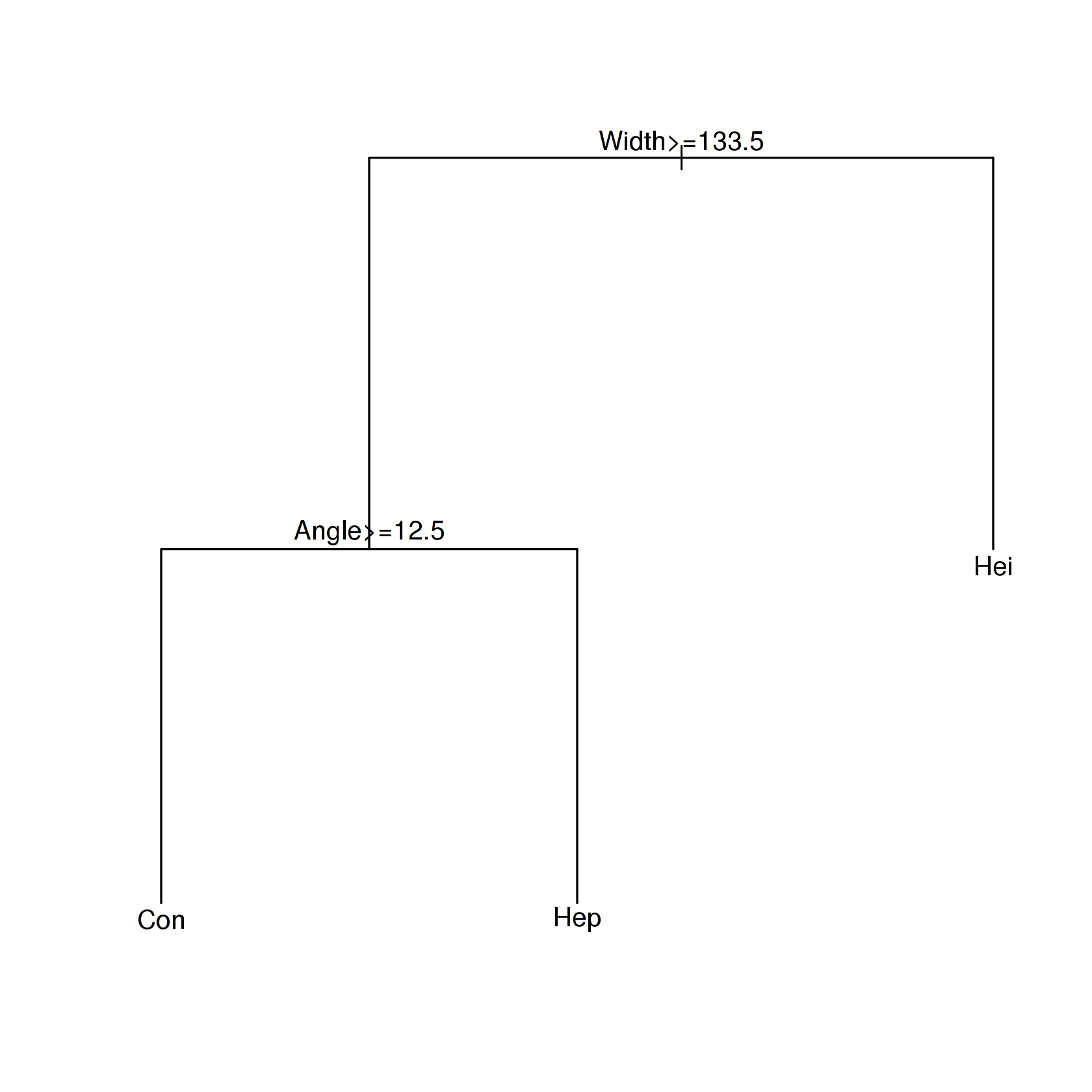
Деревья решений
Loading required package: grid
Loading required package: mvtnorm
Loading required package: modeltools
Loading required package: stats4
Loading required package: strucchange
Loading required package: zoo
Attaching package: ‘zoo’
The following objects are masked from ‘package:base’:
as.Date, as.Date.numeric
Loading required package: sandwich
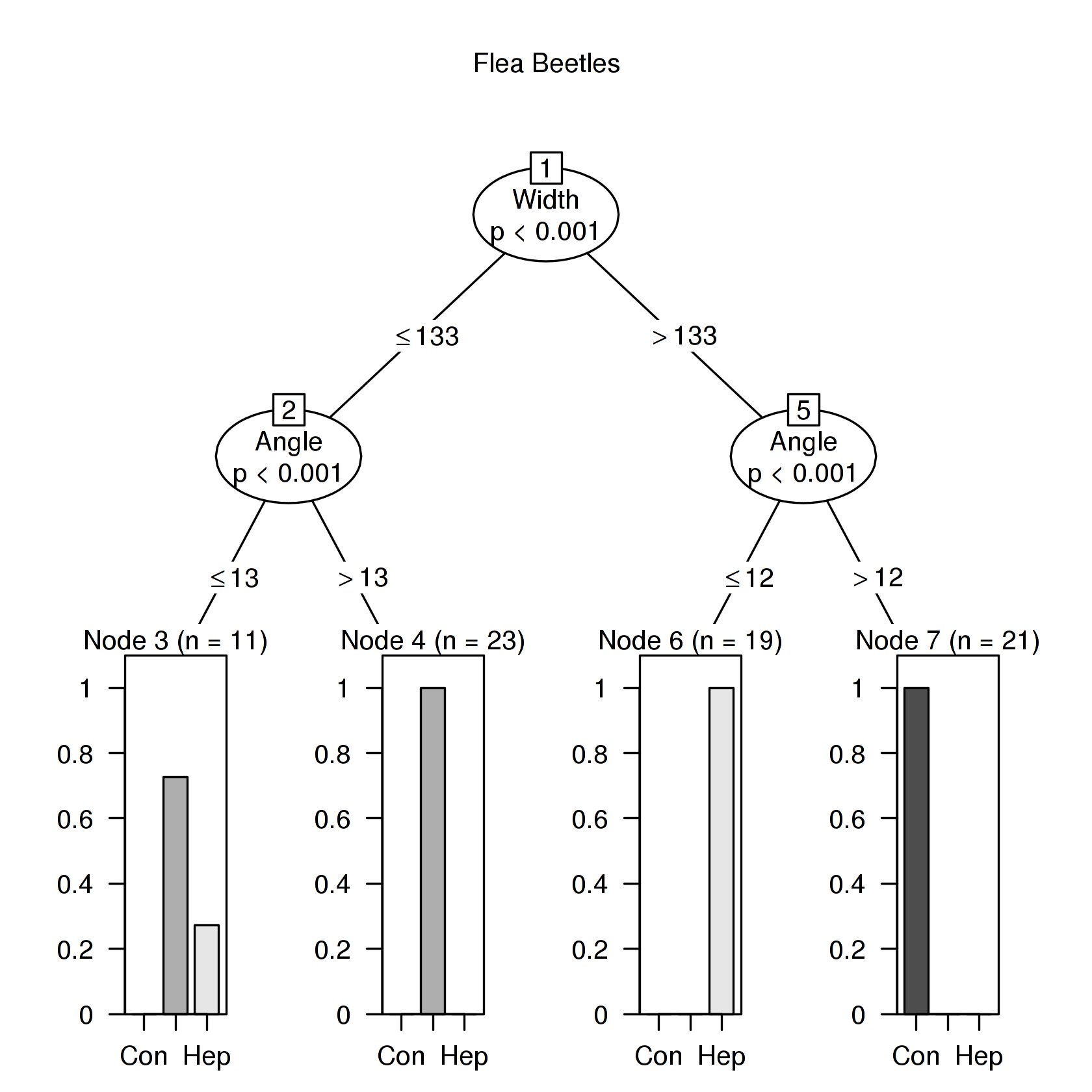
Random Forest
Call:
randomForest(formula = Species ~ ., data = flea)
Type of random forest: classification
Number of trees: 500
No. of variables tried at each split: 1
OOB estimate of error rate: 2.7%
Confusion matrix:
Con Hei Hep class.error
Con 20 1 0 0.04761905
Hei 0 30 1 0.03225806
Hep 0 0 22 0.00000000
| MeanDecreaseGini | |
|---|---|
| Width | 26.11666 |
| Angle | 20.74728 |
Предсказание
- 3
- Con
- 4
- Con
- 6
- Con
- 7
- Con
- 9
- Con
- 11
- Con
- 17
- Hei
- 19
- Con
- 20
- Con
- 25
- Hei
- 31
- Hei
- 40
- Hei
- 42
- Hei
- 43
- Hei
- 49
- Hei
- 50
- Hei
- 53
- Hep
- 57
- Hep
- 61
- Hep
- 62
- Hep
- 63
- Hep
- 65
- Hep
- 69
- Hep
- 74
- Hep
flea.rfp Con Hei Hep
Con 8 0 0
Hei 1 7 0
Hep 0 0 8
Визуализация